CompassDB: a data warehouse for single-cell Multiome (co-assay of RNA+ATAC) data
Overview
CompassDB is a data warehouse designed for single-cell multi-omics data. Specifically, it collects public single-cell RNA-Seq and single-cell ATAC-seq co-assay data from GEO and ENCODE. We process this data using a uniform data processing pipeline to prevent biases introduced by different processing methods. Additionally, we have developed a user-friendly interface for researchers.
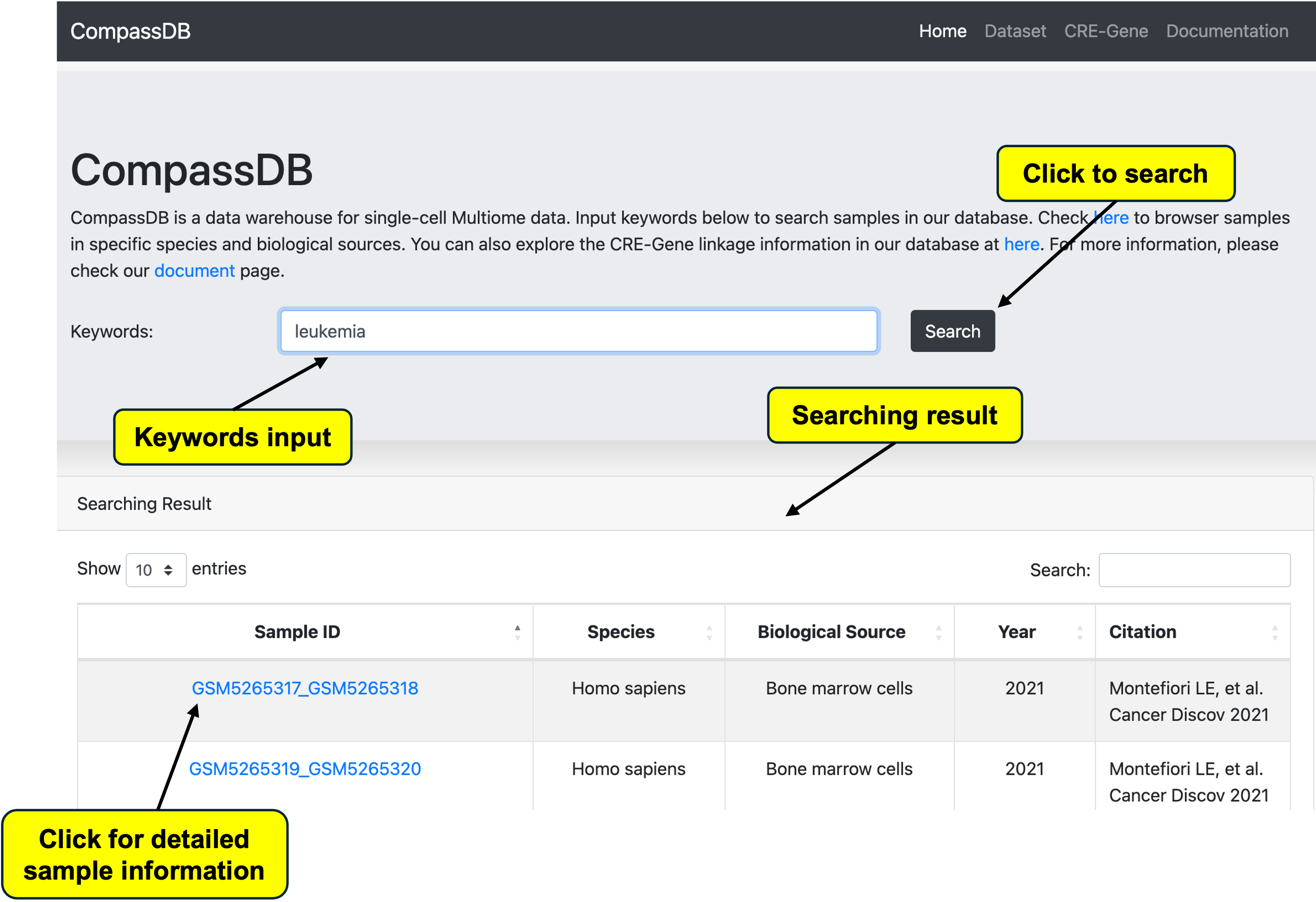
Search by Keywords
Users can search our database using keywords such as species, tissue, GSEM ID, and more. After entering keywords and clicking the search button, the result panel will display with brief targeted sample information. For detailed sample information, users can click the ID link to be redirected to the sample information page.
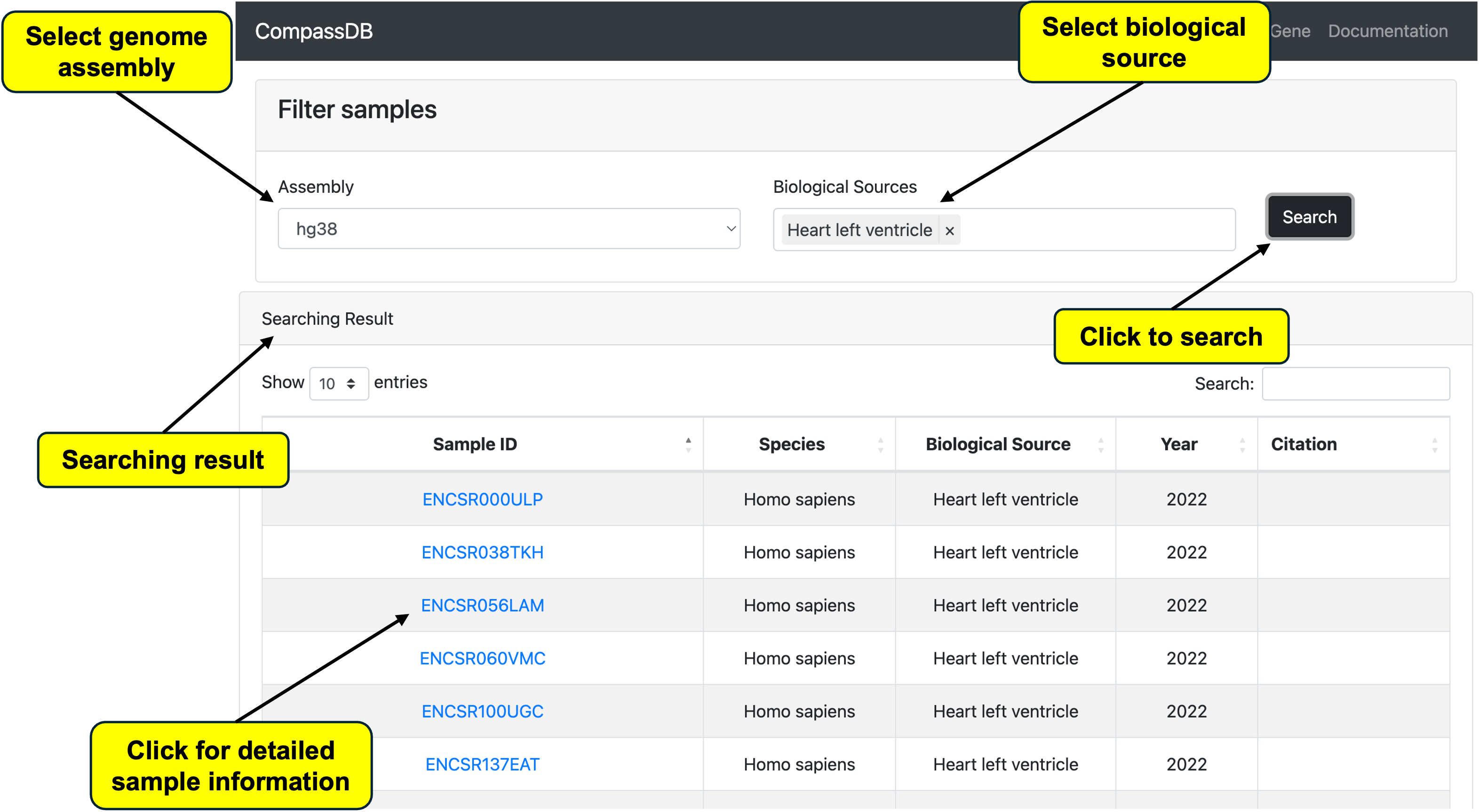
Search by Tissues
Most researchers are interested in samples from specific biological systems. We have also developed a function to search for samples based on biological sources (e.g., tissues). After selecting an assembly box, the tissue input box updates automatically. Users can select one or multiple tissues.
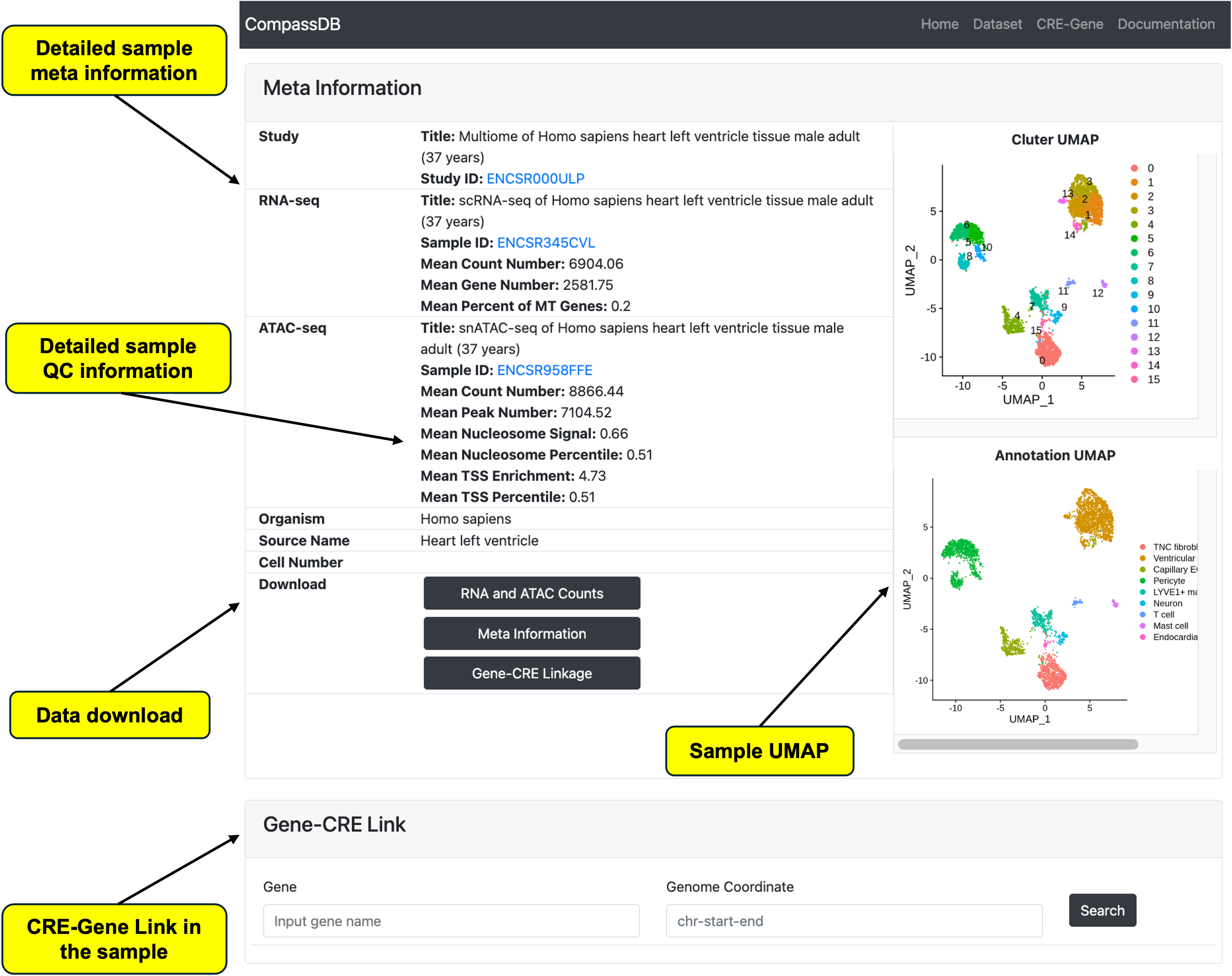
Sample Information Page
As mentioned above, users can click on a sample ID to access the detailed sample information page. On this page, researchers can review the sample metadata, including publication information and the original data link. The page also includes quality control metrics to help users decide whether to include the sample in their study. For those who require higher-level data reuse and need the original data, there is a download function to obtain the original count matrix, metadata (including QC information), and CRE-gene linkage. Additionally, the page features a tool panel to search for gene and CRE linkage within the sample.
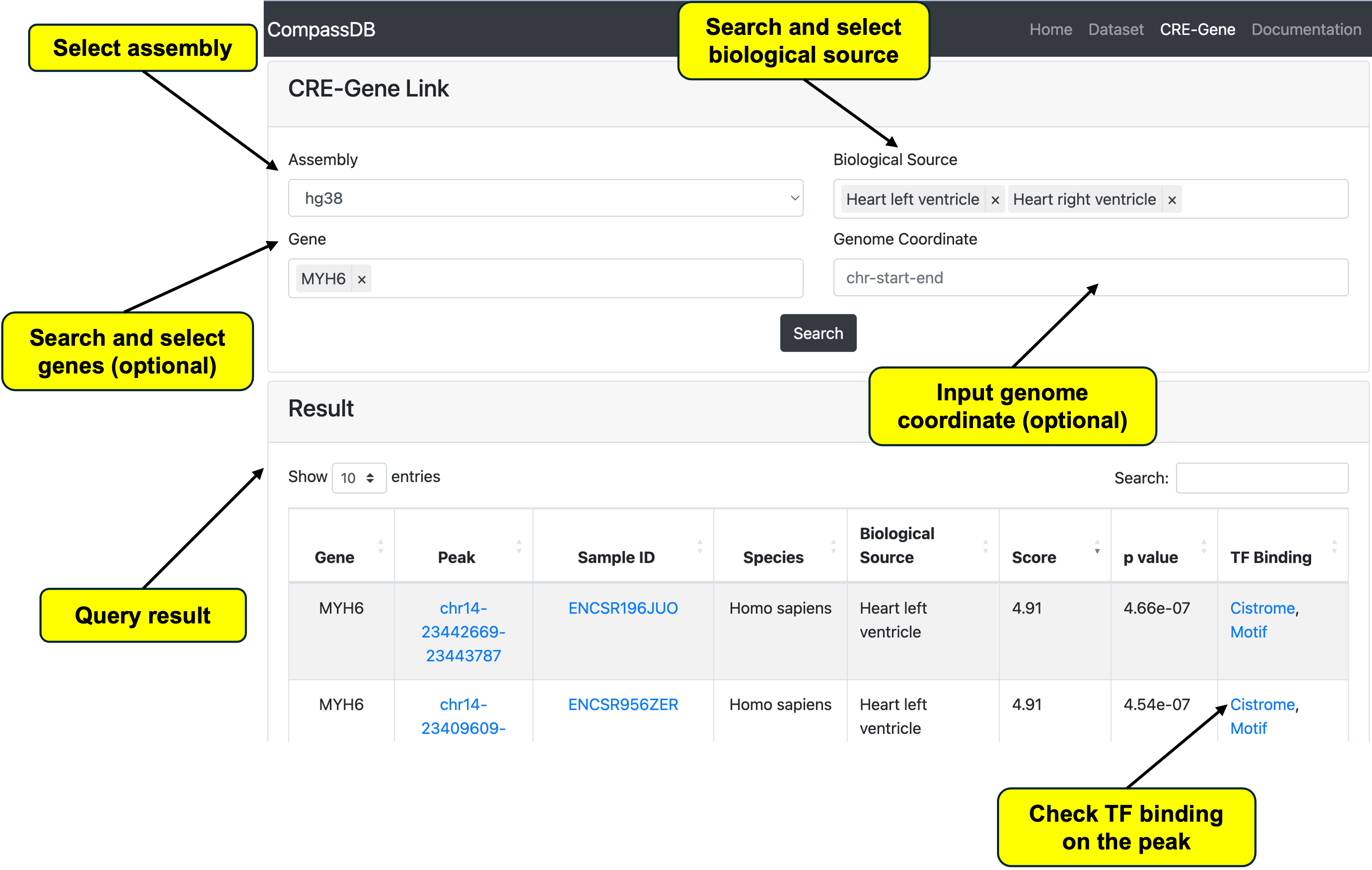
Search CRE-gene Link
Since single-cell multi-omics data can perform both RNA-seq and ATAC-seq in a single cell, it has been widely applied to study gene transcription regulation. We use the LinkPeaks function from Signac to link CREs to their targeted genes. We also provide a function to query all CRE-gene linkages in our database, available under the CRE-Gene page. Users must specify the assembly selection, while the biological source, gene, and genome coordinate are optional. For genome coordinates, users can input a region of interest (e.g., a specific transcription factor binding site), and the system will query all overlapping CREs from the database.
Browser Compatibility
| OS | Version | Chrome | Firefox | Safari | Microsoft Edge |
|---|---|---|---|---|---|
| MacOS | 12.4 | 103.0.5060.53 | 64.0 | 10.1.2 | n/a |
| Linux | ubuntu 18.04.1 LTS | 71.0.3578.98 | 61.0.1 | n/a | n/a |
| Windows | 10 | 72.0.3610.2 | 64.0 | n/a | 44.177763.1.0 |
Implementation
A detailed description of the implementation: CompassDB's implementation is divided into two parts: back-end and front-end (interface). The back-end MySQL database queries the input parameters from the front-end and sends the results out using an API. The Django framework is used to build the back-end of the CompassDB server. For the front-end, HTML and JavaScript are the main programming languages.
FAQ
1. Which genome assemblies can be used by CompassDB?
Currently, CompassDB only supports two species (human and mouse) and two genome assembly versions for each species (hg38 for human; mm10 for mouse).
2. How to cite CompassDB?
We are preparing manuscript. People should also cite the original publication if they use a single sample.